
High proportion of wild-type gastrointestinal stromal tumor in a cohort of Chilean patients screened by KIT and PDGFRA exome profiling
Introduction
Although gastrointestinal stromal tumors (GIST) are rare, they are the most frequent mesenchymal malignancies of the GI tract with an estimated incidence of 1.5 per 100,000/year (1). Studies suggest these tumors may arise or differentiate from interstitial Cajal cells or their precursor stem cells (2). The most frequent primary site is the stomach (45–59%), followed by the small intestine (30–45%) (1,3,4). In terms of survival, GIST patients usually display prolonged overall survival (OS) rates ranging from 8.9 to 13 years. Similar to many cancer types, the incidence and prevalence of GIST is geographically heterogeneous. Recently, we reported that the proportion of metastatic patients displayed significant differences when comparing Chilean and Mexican GIST registries (5).
Several reports have indicated that KIT proto-oncogene (KIT) and platelet-derived growth factor alpha (PDGFRA) mutations are very common in GISTs, affecting 80–90% of patients (6). Indeed, given its high mutation rate, KIT serves both as a hallmark for GISTs and as an actionable target. Imatinib is a tyrosine kinase inhibitor (TKI) originally designed to target the BCR-ABL fusion kinase in the late 1990s; however, subsequent studies revealed that imatinib also can also target KIT and PDGFRA. Consequently, in 2002, imatinib mesylate became the standard of care for advanced stage GISTs (7,8). Studies have confirmed the efficacy of imatinib, demonstrating up to 13% of disease control in metastatic or unresectable patients even after 10 years (9). Previous reports have also demonstrated that KIT and PDGFRA mutant patients experience the most benefits from imatinib treatment (10,11). Despite this, a proportion of patients are refractory to imatinib treatment including exon 9 KIT mutants and KIT/PDGFRA wild-types (10,12). Moreover, a subset of KIT-mutants are characterized by good initial responses to imatinib, but later these patients become refractory (12). The therapeutic alternative for these patients is sunitinib malate, a broad-spectrum TKI that also exhibits anti-angiogenic properties (13). Interestingly, a study demonstrates that some of these patients harbor pathogenic KIT mutations and are therefore eligible for KIT-specific therapies (14).
In addition to first-line imatinib and sunitinib, other TKIs, such as larotrectinib and entrectinib, are recommended for GIST patients that display NTRK fusions (15). Others, such as avapritinib, are specifically designed for PDGFRA mutations, such as D842V on exon 18 or ripretinib for KIT/PDGFRA mutations that affect exons 9, 11, 13, 14, 17 or 18, including D842V (3,16,17). Therefore, the identification of primary/secondary KIT/PDGFRA mutations provides key therapeutic information for these treatments, and could identify alternative approaches in anticipation of the development of drug resistance in GIST patients (18-20).
Here, we report the mutational profiling of KIT/PDGFRA in a group of 42 Chilean GIST patients, describing their main characteristics and specific alterations. Expectedly, our analysis found that most patients benefited from first-line imatinib or sunitinib treatment. We hypothesized that complementary analyses that encompassed all KIT and PDGFRA genes could expand this benefit to a particular subset of patients categorized as wild-type (WT) due to the absence of alterations in specific KIT/PDGFRA exons. As a proof of principle, we present the case of an advanced stage GIST patient initially classified as WT and analyzed with next-generation sequencing (NGS). In view of our results that demonstrate the clinical utility of precision medicine tools (such as NGS), we support the use of NGS for metastatic or recurrent advanced stage GISTs. We present the following article in accordance with the STROBE reporting checklist (available at https://gist.amegroups.com/article/view/10.21037/gist-21-19/rc).
Methods
Patients
In this observational study, we included the KIT/PDGFRA mutational status reports of 42 GIST patients of the Centro de Cancer UC-CHRISTUS of the Pontificia Universidad Catolica de Chile (PUC), who had been diagnosed with GIST from February 2002 to January 2019. These patients underwent KIT/PDGFRA mutational profiling during their diagnosis or treatment, with molecular testing results from November 2015 to February 2019. All mutational status reports were considered for the study size. For this study, patients were registered between May 2017 and December 2019 in a patient registry led by “Fundacion GIST Chile”, a patient advocacy group dedicated to accompanying and educating GIST patients, developing public policies to include GISTs and their treatments to the Chilean public health system, and contributing to research efforts studying this entity. Patients were included if their data was available from clinical records, the had a confirmed histopathologic diagnosis of GIST, and they were ≥18 years old. Patients with missing or incomplete clinical records, including follow-up data, or who were unable to read and write an informed consent form were excluded from this study.
Recorded variables of interest included KIT/PDGFRA alteration per exon, patient gender, age at diagnosis, disease stage at diagnosis, primary tumor location, mitotic index, tumor size, and the modified National Institutes of Health (NIH) risk of recurrence. These data were obtained from medical records and available reports. Quantitative data were grouped and handled by intervals.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Internal Review Board and the Ethics and Scientific Committee of the Pontificia Universidad Católica de Chile (approval #16-046) and individual consent for this retrospective analysis was waived.
Molecular analysis
Formalin-fixed paraffin-embedded (FFPE) tissue samples from primary or secondary GISTs were obtained from all 42 participants. Hematoxylin-eosin staining and light microscopy inspection were used to recognize areas abundant with tumor cells. These areas were dissected from the FFPE blocks for the enrichment of cellular subpopulations. Later, DNA was extracted with the QiAMP DNA Micro Kit (Qiagen, Hilden, Germany). The KIT/PDGFRA mutational status assessment was performed on the ABI PRISM 3500XL Genetic Analyzer sequencer (Thermo Fisher Scientific, Waltham, MA, USA). The procedure involved DNA amplification by polymerase chain reaction (PCR), followed by bidirectional sequencing of KIT gene exons 9, 11, 13, and 17 and PDGFRA gene exons 12, 14, and 18. Comprehensive NGS sequencing was performed using a panel of 688 cancer-related genes (Sentis Cancer + Discovery; BGI, Shenzhen, China).
Results
Patient characteristics
A total of 42 patients enrolled between 2017 and 2019 were included to this study. The median age at diagnosis was 53 years old, and patients were predominantly female (n=22; 52.4%). As expected, the stomach was the most frequent primary tumor location (n=20; 47.6%) followed by the small intestine (n=16; 38.1%) and 16 patients (38.1%) had metastatic disease at diagnosis. Regarding the risk of recurrence, 73% (n=16) of patients with resected localized disease were classified as high risk according to the modified NIH risk stratification criteria. Clinical data, including mitotic index and tumor size are summarized in Table 1.
Table 1
| Characteristics | Study population, n (%) |
|---|---|
| Gender | |
| Male | 20 (47.6) |
| Female | 22 (52.4) |
| Age at diagnosis, years | |
| 18–40 | 6 (14.3) |
| 41–60 | 23 (54.8) |
| >60 | 13 (31.0) |
| Median (IQR) | 53 (16.8) |
| Stage at diagnosis | |
| Localized | 26 (61.9) |
| Metastatic | 16 (38.1) |
| Primary tumor location | |
| Stomach | 20 (47.6) |
| Small intestine | 16 (38.1) |
| Rectum | 5 (11.9) |
| EGIST | 1 (2.4) |
| Mitotic index | |
| ≤5/50 HPF | 16 (38.1) |
| >5/50 HPF | 18 (42.9) |
| NA | 8 (19.0) |
| Tumor size*, cm | |
| 2.0–4.9 | 7 (26.9) |
| 5.0–10.0 | 12 (46.2) |
| >10.0 | 4 (15.4) |
| NA | 3 (11.5) |
| Modified NIH risk of recurrence** | |
| Low | 0 (0.0) |
| Intermediate | 6 (27.3) |
| High | 16 (72.7) |
*, tumor size assessed in localized, surgically resected GISTs (n=26); **, modified NIH risk of recurrence assessed in localized, surgically resected GISTs with available data only (n=22). IQR, interquartile range; EGIST, extra gastrointestinal stromal tumor; HPF, high power field; NA, not available.
Molecular profile in KIT and PDGFRA genes
As expected, KIT and PDGFRA mutations were mutually exclusive (Table 2); 64.3% and 4.8% harbored KIT and PDGFRA alterations, respectively. In the case of KIT, the mutations were mainly deletions located at exon 11 (n=24; 57.1%) and exon 9 (n=3; 7.1%). No alterations were found at exons 13 or 17 (Table 2). For PDGFRA, most alterations were synonymous mutations (P567P and V824V) on exons 12 and 18, and only 2 cases harbored missense mutations (D842V) at exon 18 (Table 2). When we analyzed the subset of KIT/PDGFRA mutants (n=29), 82.8% and 10.3% harbored exon 11 and exon 9 KIT mutations, respectively; 6.9% displayed exon 18 PDGFRA mutations. Interestingly, a high percentage of patients (n=13; 31.0%) had no alterations on either KIT or PDGFRA and therefore, they could be categorized as WT GISTs. Figure 1 summarizes our findings and their clinical implications.
Table 2
| No. | KIT | PDGFRA | KIT status | PDGFRA status | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exon 9 | Exon 11 | Exon 13 | Exon 17 | Exon 12 | Exon 14 | Exon 18 | ||||
| 1 | WT | DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 2 | WT | NSA | WT | WT | WT | WT | WT | ALT | WT | |
| 3 | WT | E562_P573 DEL | WT | WT | WT | WT | V824V | ALT | WT | |
| 4 | NSA | WT | WT | WT | WT | WT | WT | ALT | WT | |
| 5 | WT | WT | WT | WT | WT | WT | V824V | WT | WT | |
| 6 | WT | V559D | WT | WT | WT | WT | WT | ALT | WT | |
| 7 | WT | K558_V560 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 8 | WT | K556-558 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 9 | WT | WT | WT | WT | WT | WT | WT | WT | WT | |
| 10 | WT | NSA | WT | WT | WT | WT | WT | ALT | WT | |
| 11 | WT | WT | WT | WT | WT | WT | V824V | WT | WT | |
| 12 | WT | V559_E561 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 13 | WT | WT | WT | WT | P567P | WT | WT | WT | WT | |
| 14 | Y503_F503 INS | WT | WT | WT | WT | WT | V824V | ALT | WT | |
| 15 | WT | WT | WT | WT | P567P | WT | WT | WT | WT | |
| 16 | WT | M552_W557 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 17 | WT | V560D | WT | WT | WT | WT | WT | ALT | WT | |
| 18 | WT | W557 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 19 | WT | W557_K558 DEL | WT | WT | P567P | WT | WT | ALT | WT | |
| 20 | WT | WT | WT | WT | WT | WT | D842V | WT | ALT | |
| 21 | WT | WT | WT | WT | WT | WT | WT | WT | WT | |
| 22 | WT | D579 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 23 | WT | W557_K558 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 24 | WT | P551_Q556 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 25 | WT | NSA | WT | WT | WT | WT | WT | ALT | WT | |
| 26 | WT | WT | WT | WT | WT | WT | D842V | WT | ALT | |
| 27 | WT | V559D | WT | WT | WT | WT | V824V | ALT | WT | |
| 28 | WT | WT | WT | WT | P567P | WT | V824V | WT | WT | |
| 29 | WT | WT | WT | WT | P567P | WT | WT | WT | WT | |
| 30 | WT | WT | WT | WT | WT | WT | WT | WT | WT | |
| 31 | WT | V559G | WT | WT | WT | WT | WT | ALT | WT | |
| 32 | WT | WT | WT | WT | WT | WT | WT | WT | WT | |
| 33 | WT | W557_K558 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 34 | WT | K550 _K558 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 35 | WT | W557R | WT | WT | WT | WT | WT | ALT | WT | |
| 36 | WT | V560G | WT | WT | P567P | WT | WT | ALT | WT | |
| 37 | WT | W557_K558 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 38 | Y503_F504 INS | WT | WT | WT | WT | WT | WT | ALT | WT | |
| 39 | WT | N564H/N566_N574 DEL | WT | WT | WT | WT | WT | ALT | WT | |
| 40 | WT | WT | WT | WT | WT | WT | WT | WT | WT | |
| 41 | WT | WT | WT | WT | WT | WT | WT | WT | WT | |
| 42 | WT | WT | WT | WT | P567P | WT | V824V | WT | WT | |
| Total n (%) | 3 (7.1) | 24 (57.1) | 0 | 0 | 0 | 0 | 2 (4.8) | 27 (64.3) | 2 (4.8) | |
P567P and V824V in PDGFRA were not considered in the mutation frequency calculation as they were not classified as pathogenic according to current evidence. GIST, gastrointestinal stromal tumor; WT, wild-type; NSA, non-specified alteration; INS, insertion; DEL, deletion; ALT, alteration present.

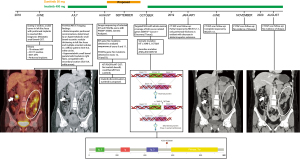
NGS in a metastatic GIST patient
Exons 9, 11, 13, 14, and 17 from KIT and exons 12, 14, and 18 from PDGFRA were analyzed by Sanger sequencing to determine the mutational status of GIST patients. Given the high proportion of WT GISTs in our cohort we decided to perform NGS in a metastatic GIST patient to search for alterations that were not detected by our initial KIT/PDGFRA exome profiling. This patient was initially classified as KIT/PDGFRA WT by the Sanger method. In general, WT patients are usually refractory to imatinib/sunitinib treatment. Figure 2 summarizes the case. The patient was a 35-year-old pregnant female diagnosed with metastatic small bowel GIST during her cesarean delivery. Due to its aggressiveness and the early onset, a comprehensive molecular test was ordered. Our analyses with NGS found 2 clinically relevant alterations: a germline pathogenic alteration in ATM (K3016Sfs*43) and a KIT deletion (c.1648-3_1673del). The latter was a 1–14 bp deletion of intron 10 that resulted in loss of the acceptor splice site at exon 11 and produced a mutant isoform of the KIT protein and a deletion of 9 amino acids (550-KPMYEVQWK-558). Based on the reports, the patient received imatinib and has remained under this treatment for >10 months. Follow-up computed tomography (CT) scans demonstrated a sustained complete response assessed by Response Evaluation Criteria in Solid Tumors (RECIST) v1.1.
Discussion
Recently, our research group reported clinical and pathological characteristics on >600 GIST patients from Chile and Mexico, revealing geographical differences in terms of stage at diagnosis, primary tumor location and tumor size (5). Herein, we expanded on these findings and delivered a KIT/PDGFRA mutational profile of Chilean GIST patients. Overall, 69% of patients in our series displayed KIT and/or PDGFRA alterations. In particular, 64.3% and 4.8% carried KIT and PDGFRA mutations, respectively (Table 2). In contrast, clinical trials have shown that KIT and PDGFRA mutation rates range between 80% and 85% and 5% and 10%, respectively (21,22). These differences could be attributed to the characteristics of the patients in each case; while clinical trials included mostly advanced stage patients, a high proportion of patients in our study had localized GISTs (61.9%, Table 1). Previous population-based studies have reported similar discrepancies. A study by Braggio et al. found 74.5% and 7.3% of KIT and PDGFRA mutants in a Brazilian cohort of GIST patients (23). Similarly, studies based on Greek (22), Italian (24), and Chinese (25) cohorts have reported important differences in the percentages of KIT and PDGFRA mutants. Notably, we observed lower mutation rates for both PDGFRA and KIT in our cohort, and consequently a high proportion of wild types (31%, Table 2). In this regard, epidemiological studies from Northern Norway (26) and the North American Intergroup (27) found that approximately 15% of GISTs are classified as WT. This discrepancy could be due to ethnic and/or genetic variations in our study population. Genetic studies (28) have already reported the underrepresentation of certain populations which makes a fair comparison difficult. The discrepancy could also be due to differences in technical or methodological approaches. The number of alterations that could be effectively detected by the Sanger method used in our study was much lower compared to massively parallel sequencing techniques such as NGS; therefore, this methodologic bias could underestimate the true mutation rate of these genes (14). Alternately, according to the 1000-G Project, silent PDGFRA alterations such as P567P and V824V (rs1873778 and rs2228230) are considered single nucleotide polymorphisms (germline) with an allelic frequency of 1% and 28% in the American population, respectively (29). Within our cohort, 7 out of 42 patients harbored these variants. Unfortunately, since our analyses were performed on tumor tissues, we cannot ensure the germline status of these alterations. Despite this, the high frequency of the PDGFRA rs1873778 variant in our cohort warrants further investigation, particularly regarding its association with GIST risk in the Chilean population. Furthermore, silent mutations can also affect protein folding, potentially affecting their function (30,31). However, the functional impact of these highly prevalent variants is yet to be determined.
The high proportion of WT GISTs in our cohort also warrants further investigation. From a biological perspective, these cases represent a distinctive phenotype, of which evidence has demonstrated an association with TKI resistance (32). Counterintuitively, WT GISTs are not free of molecular alteration, studies have shown that a proportion of these cases are characterized by succinate dehydrogenase (SDH) deficiencies (33) or by increases in growth factor expression such as VEGF or IGFR1 (34,35). Another subset included SDH-competent cases that displayed NF1, BRAF, NRAS, and PIK3CA mutations and, less frequently, NTRK3 or FGFR1 fusions, all with important pharmacological implications (32,35).
In addition to their role as first-line systemic treatment in advanced stage or recurrent GISTs, imatinib and sunitinib can also be used as adjuvant therapy in patients with a medium/high risk of postoperative recurrence. The clinical benefit of these drugs is determined by the type and location of KIT and PDGFRA mutations, as shown in Figure 1: exon 11 KIT mutations are the most prevalent with a 70% frequency (36), and W557_K558del deletions are associated with more aggressive tumors compared to single nucleotide variation (SNV) type mutations (10,37). Nevertheless, several studies have confirmed the sensitivity to imatinib on exon 11 mutants (19,35,38). Regarding exon 9 KIT mutations, meta-analyses have shown poorer OS and response rates to imatinib compared to exon 11 mutants (39). However, studies demonstrate better OS, progression-free survival (PFS), and clinical response in these patients using higher imatinib doses (800 mg) (40). Functionally, the kinase domain of KIT is divided into 2 cluster regions: the adenosine thiphosphate (ATP)-binding pocket (spanning exons 13 and 14) and the activation loop (exons 17 and 18), and both these regions are associated with secondary mutations that confer imatinib resistance (41). It is common for PDGFRA mutations to be found in the exon 18 region. The most common mutation in this exon is p.D842V, a variant associated with imatinib resistance (38). In contrast, exon 12 and exon 14 PDGFRA mutations are typically rare, and in most cases maintain imatinib responsiveness (42).
As explained, our technical and methodological approach could have underestimated the proportion of mutant GISTs or led to misinterpretations regarding their true prevalence in our cohort. The patient presented in Figure 2 is a case in point. This patient was originally diagnosed with metastatic GIST. Initially, the main exons of KIT and PDGFRA were genotyped using the Sanger method as a part of the routine procedure for GIST patients in our health center. Hence, these studies concluded the patient was a WT (no mutations). However, subsequent NGS analyses found an intronic KIT mutation close to the intron 10-exon 11 junction. This alteration has been previously reported and categorized as being sensitive to imatinib. Under treatment with imatinib the patient showed no evidence of disease for 10 months (Figure 2). Some important lessons can be learned from this case. First, as recommended by Corless et al., laboratories that usually screen for KIT mutations should anticipate intron-10-exon 11-boundary mutations, these are not uncommon and may be overlooked when PCR primers are too proximal to the exon 11 using the Sanger method (43). Second, deep genomic profiling should be considered as an option especially for advanced stage patients given its potential clinical utility (44). Within this context, precision oncology might offer substantial benefits in terms of treatment, particularly on advanced stage or refractory GIST patients. Finally, our work had several limitations. First, this was a retrospective study that reported on the data of a relatively small sample of patients. Second, the access to clinical data was limited and therefore we did not include survival outcomes, such as OS or PFS, or information on the response to treatment or its association with molecular alterations.
Conclusions
We have reported an unexpectedly high proportion of WT GIST in our Chilean cohort. These results could be attributed to technical/methodological and/or ethnic/genetic differences. In view of these findings, we support the use of alternative/complementary methods, such as NGS, for the screening of KIT and PDGFRA mutations, especially on advanced stage patients. These approaches could also reveal therapeutic alternatives for hard-to-treat patients. Future studies should determine if this is a feasible, cost-effective strategy.
Acknowledgments
We thank the study participants, the University Hospital medical and nursing staff, Fundación GIST Chile, and Pfizer for their contribution to the patients and this study.
Funding: This work was supported by educational support for non-profit organizations by Pfizer Inc.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://gist.amegroups.com/article/view/10.21037/gist-21-19/rc
Data Sharing Statement: Available at https://gist.amegroups.com/article/view/10.21037/gist-21-19/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gist.amegroups.com/article/view/10.21037/gist-21-19/coif). PF has received financial support from Pfizer Inc. and Roche. MG has been a member of Scientific Advisory Boards during the past 36 months at Bayer, Novartis, MSD, BMS, Pfizer, Macrogenic, and Merck, has participated as an invited speaker in activities by Novartis, Pfizer, Bayer, BMS, MSD, GBT Biotoscana, and Lilly and has received traveling accommodation and attending support for international meetings by Novartis, MSD, and Bayer. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Internal Review Board and the Ethics and Scientific Committee of the Pontificia Universidad Católica de Chile (approval #16-046) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Søreide K, Sandvik OM, Søreide JA, et al. Global epidemiology of gastrointestinal stromal tumours (GIST): A systematic review of population-based cohort studies. Cancer Epidemiol 2016;40:39-46. [Crossref] [PubMed]
- Min KW, Leabu M. Interstitial cells of Cajal (ICC) and gastrointestinal stromal tumor (GIST): facts, speculations, and myths. J Cell Mol Med 2006;10:995-1013. [Crossref] [PubMed]
- IJzerman NS, Drabbe C, den Hollander D, et al. Gastrointestinal Stromal Tumours (GIST) in Young Adult (18-40 Years) Patients: A Report from the Dutch GIST Registry. Cancers (Basel) 2020;12:730. [Crossref] [PubMed]
- Shen C, Wang C, He T, et al. Long-term survival among patients with gastrointestinal stromal tumors diagnosed after another malignancy: a SEER population-based study. World J Surg Oncol 2020;18:88. [Crossref] [PubMed]
- Calderillo G, Muñoz-Medel M, Carbajal E, et al. Retrospective Analysis of Chilean and Mexican GI Stromal Tumor Registries: A Tale of Two Latin American Realities. JCO Glob Oncol 2020;6:647-57. [Crossref] [PubMed]
- Kelly CM, Gutierrez Sainz L, Chi P. The management of metastatic GIST: current standard and investigational therapeutics. J Hematol Oncol 2021;14:2. [Crossref] [PubMed]
- Reichardt P. The Story of Imatinib in GIST - a Journey through the Development of a Targeted Therapy. Oncol Res Treat 2018;41:472-7. [Crossref] [PubMed]
- Dagher R, Cohen M, Williams G, et al. Approval summary: imatinib mesylate in the treatment of metastatic and/or unresectable malignant gastrointestinal stromal tumors. Clin Cancer Res 2002;8:3034-8. [PubMed]
- Casali PG, Zalcberg J, Le Cesne A, et al. Ten-Year Progression-Free and Overall Survival in Patients With Unresectable or Metastatic GI Stromal Tumors: Long-Term Analysis of the European Organisation for Research and Treatment of Cancer, Italian Sarcoma Group, and Australasian Gastrointestinal Trials Group Intergroup Phase III Randomized Trial on Imatinib at Two Dose Levels. J Clin Oncol 2017;35:1713-20. [Crossref] [PubMed]
- Joensuu H, Wardelmann E, Sihto H, et al. Effect of KIT and PDGFRA Mutations on Survival in Patients With Gastrointestinal Stromal Tumors Treated With Adjuvant Imatinib: An Exploratory Analysis of a Randomized Clinical Trial. JAMA Oncol 2017;3:602-9. [Crossref] [PubMed]
- Joensuu H, Eriksson M, Sundby Hall K, et al. Survival Outcomes Associated With 3 Years vs 1 Year of Adjuvant Imatinib for Patients With High-Risk Gastrointestinal Stromal Tumors: An Analysis of a Randomized Clinical Trial After 10-Year Follow-up. JAMA Oncol 2020;6:1241-6. [Crossref] [PubMed]
- Dematteo RP, Ballman KV, Antonescu CR, et al. Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet 2009;373:1097-104. [Crossref] [PubMed]
- Hao Z, Sadek I. Sunitinib: the antiangiogenic effects and beyond. Onco Targets Ther 2016;9:5495-505. [Crossref] [PubMed]
- Astolfi A, Indio V, Nannini M, et al. Targeted Deep Sequencing Uncovers Cryptic KIT Mutations in KIT/PDGFRA/SDH/RAS-P Wild-Type GIST. Front Oncol 2020;10:504. [Crossref] [PubMed]
- Demetri GD, Antonescu CR, Bjerkehagen B, et al. Diagnosis and management of tropomyosin receptor kinase (TRK) fusion sarcomas: expert recommendations from the World Sarcoma Network. Ann Oncol 2020;31:1506-17. [Crossref] [PubMed]
- Avapritinib Approved for GIST Subgroup. Cancer Discov 2020;10:334. [PubMed]
- Blay JY, Serrano C, Heinrich MC, et al. Ripretinib in patients with advanced gastrointestinal stromal tumours (INVICTUS): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2020;21:923-34. [Crossref] [PubMed]
- Ravegnini G, Nannini M, Sammarini G, et al. Personalized Medicine in Gastrointestinal Stromal Tumor (GIST): Clinical Implications of the Somatic and Germline DNA Analysis. Int J Mol Sci 2015;16:15592-608. [Crossref] [PubMed]
- Tan S, Chen P, Ji J, et al. Genomic Subtypes of GISTs for Stratifying Patient Response to Sunitinib following Imatinib Resistance: A Pooled Analysis and Systematic Review. Dis Markers 2018;2018:1368617. [Crossref] [PubMed]
- Wang Y, Call J. Mutational Testing in Gastrointestinal Stromal Tumor. Curr Cancer Drug Targets 2019;19:688-97. [Crossref] [PubMed]
- Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 2003;21:4342-9. [Crossref] [PubMed]
- Mavroeidis L, Metaxa-Mariatou V, Papoudou-Bai A, et al. Comprehensive molecular screening by next generation sequencing reveals a distinctive mutational profile of KIT/PDGFRA genes and novel genomic alterations: results from a 20-year cohort of patients with GIST from north-western Greece. ESMO Open 2018;3:e000335. [Crossref] [PubMed]
- Braggio E, Braggio Dde A, Small IA, et al. Prognostic relevance of KIT and PDGFRA mutations in gastrointestinal stromal tumors. Anticancer Res 2010;30:2407-14. [PubMed]
- Braconi C, Bracci R, Bearzi I, et al. KIT and PDGFRalpha mutations in 104 patients with gastrointestinal stromal tumors (GISTs): a population-based study. Ann Oncol 2008;19:706-10. [Crossref] [PubMed]
- Du CY, Shi YQ, Zhou Y, et al. The analysis of status and clinical implication of KIT and PDGFRA mutations in gastrointestinal stromal tumor (GIST). J Surg Oncol 2008;98:175-8. [Crossref] [PubMed]
- Steigen SE, Eide TJ, Wasag B, et al. Mutations in gastrointestinal stromal tumors--a population-based study from Northern Norway. APMIS 2007;115:289-98. [Crossref] [PubMed]
- Heinrich MC, Owzar K, Corless CL, et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup Phase III Trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 Study by Cancer and Leukemia Group B and Southwest Oncology Group. J Clin Oncol 2008;26:5360-7. [Crossref] [PubMed]
- Nugent A, Conatser KR, Turner LL, et al. Reporting of race in genome and exome sequencing studies of cancer: a scoping review of the literature. Genet Med 2019;21:2676-80. [Crossref] [PubMed]
- 1000 Genomes Project Consortium; Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015;526:68-74.
- Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science 2007;315:525-8. Erratum in: Science 2007;318:1382-3 Erratum in: Science 2011;334:39. [Crossref] [PubMed]
- Kartha RV, Sundram UN. Silent mutations in KIT and PDGFRA and coexpression of receptors with SCF and PDGFA in Merkel cell carcinoma: implications for tyrosine kinase-based tumorigenesis. Mod Pathol 2008;21:96-104. [Crossref] [PubMed]
- Wada R, Arai H, Kure S, et al. "Wild type" GIST: Clinicopathological features and clinical practice. Pathol Int 2016;66:431-7. [Crossref] [PubMed]
- Nannini M, Biasco G, Astolfi A, et al. An overview on molecular biology of KIT/PDGFRA wild type (WT) gastrointestinal stromal tumours (GIST). J Med Genet 2013;50:653-61. [Crossref] [PubMed]
- Nannini M, Astolfi A, Paterini P, et al. Expression of IGF-1 receptor in KIT/PDGF receptor-α wild-type gastrointestinal stromal tumors with succinate dehydrogenase complex dysfunction. Future Oncol 2013;9:121-6. [Crossref] [PubMed]
- Blay JY, Kang YK, Nishida T, et al. Gastrointestinal stromal tumours. Nat Rev Dis Primers 2021;7:22. [Crossref] [PubMed]
- Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol 2004;22:3813-25. [Crossref] [PubMed]
- Wozniak A, Rutkowski P, Schöffski P, et al. Tumor genotype is an independent prognostic factor in primary gastrointestinal stromal tumors of gastric origin: a european multicenter analysis based on ConticaGIST. Clin Cancer Res 2014;20:6105-16. [Crossref] [PubMed]
- Zhang H, Liu Q. Prognostic Indicators for Gastrointestinal Stromal Tumors: A Review. Transl Oncol 2020;13:100812. [Crossref] [PubMed]
- Yan L, Zou L, Zhao W, et al. Clinicopathological significance of c-KIT mutation in gastrointestinal stromal tumors: a systematic review and meta-analysis. Sci Rep 2015;5:13718. [Crossref] [PubMed]
- Gastrointestinal Stromal Tumor Meta-Analysis Group (MetaGIST). Comparison of two doses of imatinib for the treatment of unresectable or metastatic gastrointestinal stromal tumors: a meta-analysis of 1,640 patients. J Clin Oncol 2010;28:1247-53. [Crossref] [PubMed]
- Serrano C, Mariño-Enríquez A, Tao DL, et al. Complementary activity of tyrosine kinase inhibitors against secondary kit mutations in imatinib-resistant gastrointestinal stromal tumours. Br J Cancer 2019;120:612-20. [Crossref] [PubMed]
- Lasota J, Stachura J, Miettinen M. GISTs with PDGFRA exon 14 mutations represent subset of clinically favorable gastric tumors with epithelioid morphology. Lab Invest 2006;86:94-100. [Crossref] [PubMed]
- Corless CL, McGreevey L, Town A, et al. KIT gene deletions at the intron 10-exon 11 boundary in GI stromal tumors. J Mol Diagn 2004;6:366-70. [Crossref] [PubMed]
- Cobain EF, Wu YM, Vats P, et al. Assessment of Clinical Benefit of Integrative Genomic Profiling in Advanced Solid Tumors. JAMA Oncol 2021;7:525-33. [Crossref] [PubMed]
Cite this article as: Muñoz-Medel M, Córdova-Delgado M, Retamal IN, Villalobos F, Mellado R, Fernández P, Manque P, Berkovits A, Ríos JA, García-Bloj B, Rodríguez MP, Garrido M. High proportion of wild-type gastrointestinal stromal tumor in a cohort of Chilean patients screened by KIT and PDGFRA exome profiling. Gastrointest Stromal Tumor 2022;5:6.


